research
As a Bioinformatician, my research revolves around developing computational methods that can handle large amounts of complex biological data. One of the areas that I have explored extensively is alternative splicing (AS) – a complex process that plays a critical role in creating protein diversity in cells. By studying AS, we can better understand the biological mechanisms that drive it and how it contributes to various diseases.
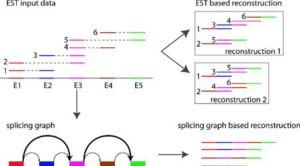
To make sense of the vast amounts of data generated by AS studies, I developed the splicing graph – an innovative graph-based representation of splice variants. With the splicing graph, we can more easily visualize and explore the relationships between different splice variants, allowing us to uncover new insights into this complex process.
Currently, I’m developing algorithms for ribosome footprinting, a cutting-edge sequencing technology that enables us to study gene translation in cells. This technique provides a unique opportunity to learn more about how genes are expressed and regulated at the protein level. While most of my research relies on using algorithm design to investigate biological data, recently, I have started a research thrust that uses nature to inspire algorithm design. More details are given in the “Projects” section below.
Research Areas:
- Algorithms and Theory of Computation
- Artificial Intelligence and Intelligent Agents
- Data Sciences and Analytics
- Graphics, Human-Computer Interaction, & User Experience
- Scientific and High-Performance Computing
Projects
Translation and Ribosome Footprinting
Ribosome footprinting is a popular technique for studying translation and its regulation. A ribosome footprinting experiment produces a snapshot of the location and abundance of actively translating ribosomes within a cell’s transcriptome.
iboStreamR is a comprehensive Ribo-seq quality control platform in the form of an R Shiny web application.
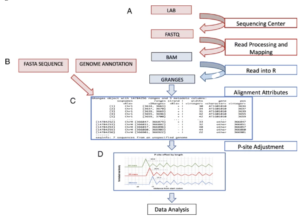
Ribosome footprinting data analysis can be sensitive to quality issues such as read length variation, low read periodicities, and contaminations with ribosomal and transfer RNA. Various software tools for data preprocessing, quality assessment, analysis, and visualization of Ribo-seq data have been developed. However, many of these tools require considerable practical knowledge of software applications, and often multiple different tools have to be used in combination with each other.RiboStreamR provides visualization and analysis tools for various Ribo-seq QC metrics, including read length distribution, read periodicity, and translational efficiency.
- Identification of Translational Hormone-Response Gene Networks and cis-Regulatory Elements. (https://www.nsf.gov/awardsearch/showAward?AWD_ID=1444561)
Traditionally scientists have studied how hormones influence gene activity by examining gene expression by measuring the copying of DNA into RNA, the first step in the transfer of genetic information into proteins, which play an enormous variety of functional roles in cells. However, an increase in expression does not always mean that more protein will be made because protein production from encoded RNA molecules is a highly regulated process. The recently developed Ribo-seq technology provides the capability of measuring protein production by every active gene in the genome at once. Genome-wide changes in translation activity in response to ethylene have been quantified at codon resolution by taking advantage of the recently developed ribosome footprinting technology. This makes it possible to identify new translational regulatory elements in Arabidopsis.
Alternative Splicing and Transcriptome Analysis
Alternative splicing is a major contributor to the diversity of the proteome, and it is involved in many animal, human, and plant diseases. One gene might produce thousands of splice variants.
The traditional approach to annotate alternative splicing is to investigate every splicing variant of the gene in a case-by-case fashion.
Splicing graphs represent each transcript by a path in the graph. Many research groups have adopted the splicing graph representation because its compact transcript representation preserves the relationships between splice variants, opening the way to investigate complex genes with thousands of splice variants.
- Transcriptional Nodes Coordinate Patterning and Cellular Proliferation During Carpel Margin Meristem Development.(https://www.nsf.gov/awardsearch/showAward?AWD_ID=1355019)
This project investigates the molecular interactions that allow seeds to develop within the flowering plant Arabidopsis thaliana. The goal is to enable future strategies to alter seed numbers in agricultural or bioenergy crops, thus supporting efforts to supply food and fuel for a growing global population. Cell sorting, in combination with a multidisciplinary integration of genetic, genomic, and bioinformatic approaches, will be applied to investigate cellular transcriptional states at an unprecedented level of specificity. - Role of alternative splicing in Arabidopsis immune response. (https://www.nsf.gov/awardsearch/showAward?AWD_ID=0951512)
Several recent reviews suggest the importance of alternative splicing in plants as a mechanism for controlling both development and stress adaptation. The goal of the proposed project is to further investigate this hypothesis by measuring the extent and functional significance of alternative splicing in Arabidopsis thaliana during its defense against the bacterial pathogen Pseudomonas syringae pv tomato.
Data-Driven Storytelling for Bioinformatics Applications
Data analysis is more than just crunching numbers and drawing charts. For communicating complex insights, you need a story that connects the dots. Data storytelling integrates data, domain expertise, intuitive visualization, and narration to create stories that are easy to understand and remember. Whether you are a scientist, a teacher, or a business professional, mastering the art of data storytelling can help you to make an impact.
- Symposium on Data Storytelling. The goal of this project is to build the foundation of an interdisciplinary research thrust that explores AI-supported natural language generation and optimized visualization for summarization and data storytelling in bioinformatics. We envision a new generation of multi-modal summarization algorithms that transform continuously increasing data volumes into engaging, user-aware, multi-modal data stories.
Infrastructure
- A Bioinformatics Computing Cluster for NC State University. (IBM Faculty Award)